Tarifs
Nous consulter.
Accueil > Plateformes > PROTEOMIC

Within the network of Paris-Saclay proteomics platforms (P2S), and affiliated to the Graduate School of Health and Drug Science (GS HeaDS), PROTEOMIC - IPSIT facility is open to researchers dedicated to pharmaceutical sciences belonging to academic or private laboratories.
PROTEOMIC - IPSIT collaborates with users by providing them technologies and expertise for qualitative and quantitative proteome analysis.

PROTEOMIC - IPSIT is listed in Pluginlabs
PRESTATIONS
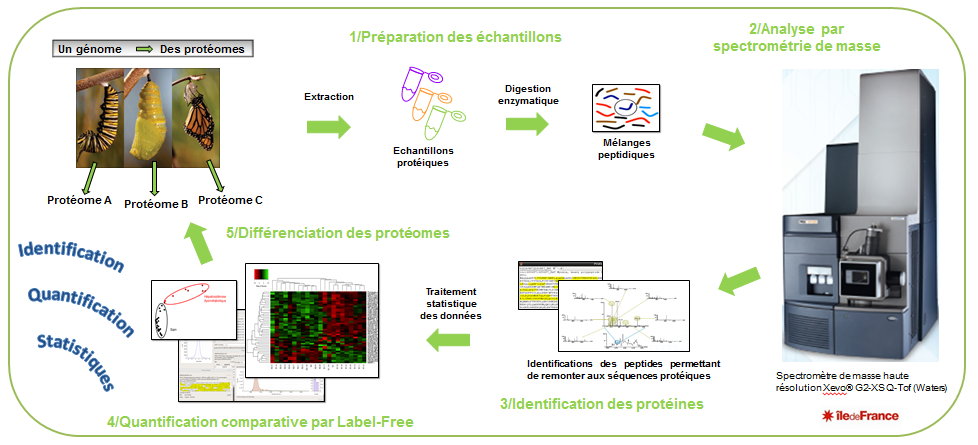
In the context of the proteome study, Bottom-Up strategy is applied. This approach involves first digesting proteins and then analyzing peptides mixture by LCMS.
The inherent complexity of the peptides mixture is resolved through nano liquid chromatography-tandem mass spectrometry analysis (LCMS). Peptides are separated under a gradient of acetonitrile on a reverse phase column. Reaching the High Resolution Mass Spectrometer, peptides are ionized by positive electrospray (ESI) and monitored depending on their mass/charge ratio (m/z). Under gas-phase collision-activated dissociation (CAD) peptides backbones are cleaved at peptide amide bonds to produce specific pattern of fragments (MS/MS).
Post-translational modifications (PTMs) can be studied, depending on their nature and stability under analysis conditions in place.
The system also allows analysis of purified intact proteins under certain conditions.
Following a meeting between the facility members and the project leader(s) to validate the feasibility of the experiments, the analyses are carried out according to the proteomics collaboration request form, filled in by mutual agreement and under conditions of collaboration described in the charter of the PROTEOMIC - IPSIT facility.
EQUIPEMENTS
Xevo G2-XS QTof - ACQUITY UPLC M-Class system (WATERS) :
Funding : Région Ile-de-France (convention EX024607), Université Paris-Saclay, INSERM and CNRS :

Bottom-Up proteomic analysis by LCMS :
The inherent complexity of the peptides mixture is resolved through nano liquid chromatography-tandem mass spectrometry analysis (LCMS). Peptides are separated under a gradient of acetonitrile on reverse phase column. Reaching the High Resolution Mass Spectrometer, peptides are ionized by positive electrospray (ESI) and monitored depending on their mass/charge ratio (m/z). Under gas-phase collision-activated dissociation (CAD) peptides backbones are cleaved at peptide amide bonds to produce specific pattern of fragments (MS/MS). Using dedicated processing softwares, proteins sequence elucidation and correlated abundance calculation are achieved.
Data Processing :
Spectrophotometer :
One-dimensional gel electrophoresis (SDS-page)
LOCALISATION ET CONTACT
stephanie.nicolay universite-paris-saclay.fr - e-mail
Tel : 01 8000 61 76
Plateforme PROTEOMIC-UMS IPSIT
UFR de Pharmacie
Université Paris-Saclay
RDC-Bâtiment Henri MOISSAN
17, Avenue des Sciences
91400 ORSAY
2020 Hélène Le Ribeuz, Florent Dumont, Guillaume Ruellou, Melanie Lambert, Thierry Balliau, Marceau Quatredeniers, Barbara Girerd, Sylvia Cohen-Kaminsky, Olaf Mercier, Stephanie Yen-Nicolaÿ, Marc Humbert, David Montani, Veronique Capuano, Fabrice Antigny. Proteomic Analysis of KCNK3 Loss of Expression Identified Dysregulated Pathways in Pulmonary Vascular Cells. Int. J. Mol. Sci. DOI : 10.3390/ijms21197400
Stéphanie YEN-NICOLAŸ Ingénieure d’études Université Paris-Saclay Responsable technique Protéomique stephanie.nicolay universite-paris-saclay.fr - e-mail Tel : 01 8000 61 76
Guillaume RUELLOU (...)